Python Read Data in Parallel
If the original data is spread out in a number of files, we can read them in parallel (on multi-core) machines to save time. Ideally. However, it seems a bit tricky with python.
Let’s assume we have a bunch of hdf5 files, containing transactional data of cryptocurrencies on Bitmex.
Presumably the data is acquired via the API provided by Bitmex.
Due to technical reasons, e.g., network interruption, the data files begin with different key appended with indicies.
I put all the hdf5 data files in a folder /data/Shared/order_book/hdf5/,
BITMEX<key>.1.h5quote
BITMEX<key>.2.h5quote
BITMEX<key>.4.h5quote
BITMEX<key>.5.h5quote
It makes sense to define some strings given the situation
data_home = '/data/Shared/order_book/hdf5/'
prefix = 'BITMEX'
postfix = 'h5quote'
product = 'XBTUSD'
All the necessary packages are imported as the follows
import os, time, math
from sys import getsizeof
from datetime import datetime, timedelta
import h5py as hdf
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
Data files integrety check
First, list all the files and identify all the files in file_list
file_list = [f for f in os.listdir(data_home) if (prefix in f) and (postfix in f)]
From file_list we get all the distinctive keys in key_list.
files = dict.fromkeys([f.split('.')[0] for f in file_list])
key_list = list(files.keys())
The files are manually uploaded. For example, if only files BITMEX<key>.1.h5quote, BITMEX<key>.2.h5quote, BITMEX<key>.4.h5quote and BITMEX<key>.5.h5quote are uploaded, we should point out that this key with index 3 is missing.
for key in files:
indicies = sorted([int(f.split('.')[1]) for f in file_list if key in f])
if indicies[len(indicies)-1]!=len(indicies)-1:
# The files are manually uploaded, with possibility of missing
print(key, " misses ",
[i for i in range(len(indicies)) if i not in indicies],
" out of ", len(indicies)-1, "files.")
# Choose not to process a key with incomplete file list.
del files[key]
else:
files[key] = indicies
Read the data sequentially
This is straightforward, with the only trouble from issues in the data files. Now, we define a function to read data.
def read_hdf5(key):
start = time.time()
try:
# some file may be broken
df[key] = pd.concat([pd.read_hdf(data_home + key + '.' + str(i) + '.' + postfix,
key=product)
for i in files[key]],
ignore_index=True)
print(key, "has",
len(df[key]), "lines of records, taking %d seconds" %
(time.time()-start))
print("Start at:", start, "end at:", datetime.fromtimestamp(time.time()))
except OSError as e:
print(e)
Now, we read the files sequentially and see how much time it takes
print("Start at:", datetime.fromtimestamp(time.time()))
df = {}
for key in files:
read_hdf5(key)
print("End at:", datetime.fromtimestamp(time.time()))
Ok, that was easy. But this takes too long to run. For about three week’s data, it takes 6 minutes using a server with 88 cores. Right, such a code does not take advantage of the multi-cores.
Start at: 2020-02-1 17:05:26.604005
BITMEX<key1> has 78786430 lines of records, taking 50 seconds
BITMEX<key2> has 7526730 lines of records, taking 46 seconds
BITMEX<key3> has 1906630 lines of records, taking 1 seconds
BITMEX<key4> has 72682736 lines of records, taking 44 seconds
BITMEX<key5> has 114284897 lines of records, taking 71 seconds
...
End at: 2020-02-1 17:11:30.964797
Parallel Processing
We will try a few different ways to do this. First, need import the following packages.
from multiprocessing import Pool, Process, Manager
With huge memory compsumption
Now, define a function to read a data file.
def read_hdf5(file):
start = time.time()
try:
# some file may be broken
df = pd.read_hdf(data_home + file, key=product)
print(file, "has",
len(df), "lines of records, taking %d seconds" %
(time.time()-start))
print(datetime.fromtimestamp(time.time()))
return file, df
except OSError as e:
print(e)
The simplest parallel reading is probably to use Pool by doing
print("Start at:", datetime.fromtimestamp(time.time()))
start = time.time()
pool = Pool(8)
df_collection = pool.map(read_hdf5, file_list)
pool.close()
pool.join()
print("End at:", datetime.fromtimestamp(time.time()))
So for each data file bitmex<key>.id.h5quote, we read it into a dataframe stored in the list df_collection.
We need to concatinate dataframes for each key according to file index order.
print("Start at:", datetime.fromtimestamp(time.time()))
df = {}
for key in files:
df_list = []
for i in files[key]:
df_list.extend([dfc[1] for dfc in df_collection if key+'.'+str(i)+'.' in dfc[0]])
df[key] = pd.concat(df_list, ignore_index=True)
print(key, "has", len(df[key]), "records.")
print("End at:", datetime.fromtimestamp(time.time()))
The above two steps cost in total 2 minutes, a three-times speed up.
Sounds good?
Yes, but this is at the cost of huge memory consumption.
As shown in htop, the memory consumption has gone from 380G to 407G just by running the parallel data reading and store all dataframe in the list df_collection.
Note that this server has 1T of memory, larger than the harddrive of mainstream desktop machines. The 27G memory consumption by this simple step is larger than most consumer level PC or Mac.
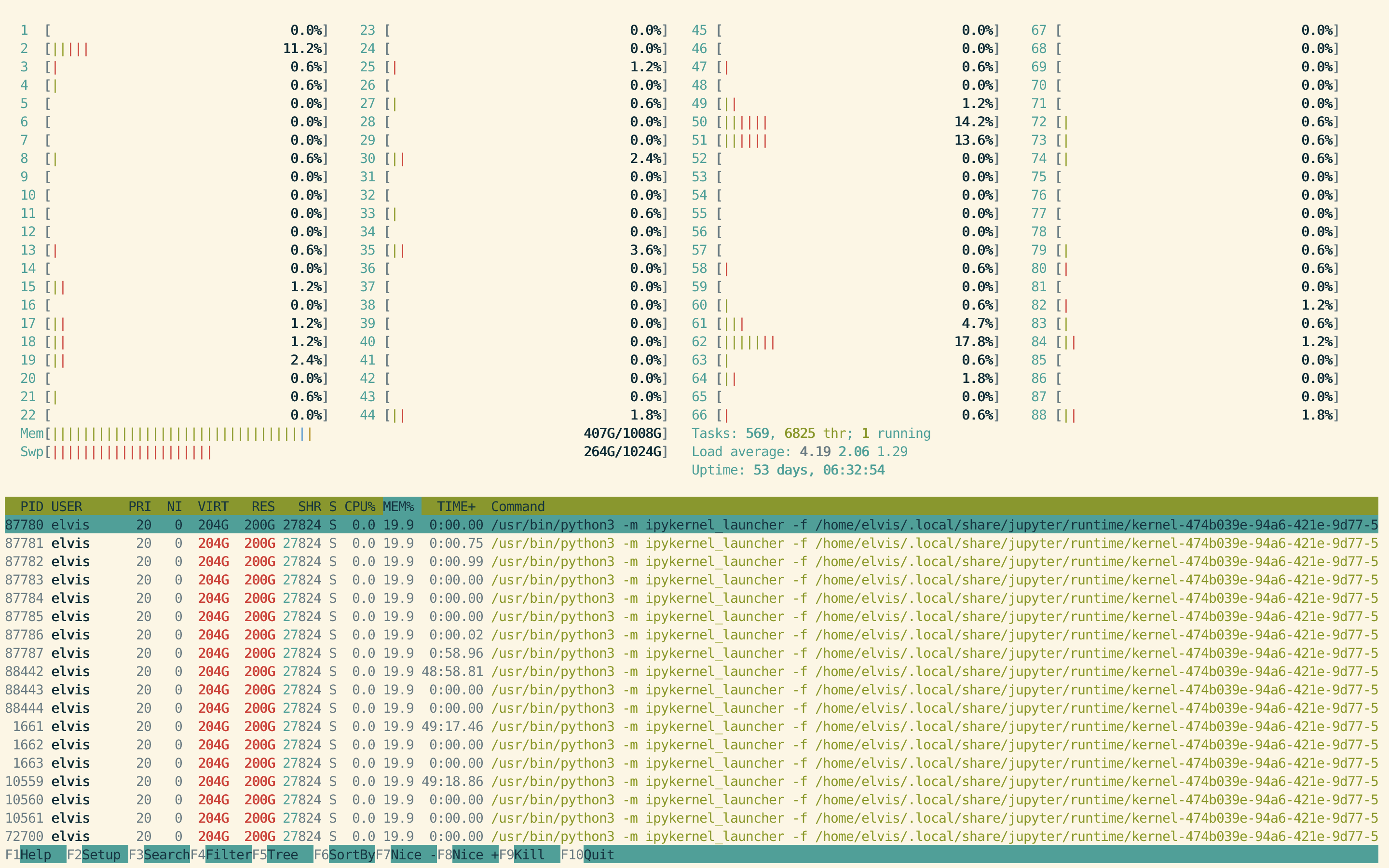
After concatinating all dataframes in df_collection accoding to each key, and storing the concatinated dataframes into df[key], an additional 35G memory is consumed.
For Python, do not expect the so-called garbage collection
import gc
gc.collect()
can help even a little bit. Neither can you expect to use del df_collection to achieve anything.
Well, if you do
for i in range(len(df_collection), 0, -1):
del(df_collection[i-1])
You will see some memory freed, roughly 15G. But remember 27G was consumed after reading files into the list of dataframes df_collection.
More on the parallel processing. You cannot naively expect to speed it up further by assigning more processes, e.g., pool = Pools(40) instead of Pool(8), since it takes time to start and close processes at least.
8 is the optimal number for this machine with 88 cores based on experiments of reading 300 data files with drastically different sizes.
Try to have a more memory-efficient solution
In Python, you can use Manager() as an agent to return valude from multiprocessing. For that, you need to redefine the readdata function.
def read_hdf5_return(key, df):
start = time.time()
try:
# some file may be broken
df[key] = pd.concat([pd.read_hdf(data_home + key + '.' + str(i) + '.' + postfix,
key=product)
for i in files[key]],
ignore_index=True)
print(key, "has",
len(df[key]), "lines of records, taking %d seconds" %
(time.time()-start))
print("Start at:", datetime.fromtimestamp(start),
"end at:", datetime.fromtimestamp(time.time()))
except OSError as e:
print(e)
Ok, now you can call the read data function with return value.
print("Start at:", datetime.fromtimestamp(time.time()))
manager = Manager()
df = manager.dict()
jobs = []
for key in files:
p = Process(target=read_hdf5_return, args=(key,df))
jobs.append(p)
p.start()
for proc in jobs:
proc.join()
print("End at:", datetime.fromtimestamp(time.time()))
It all works fine
BITMEX<key5> has 1906630 lines of records, taking 2 seconds
BITMEX<key2> has 1385198 lines of records, taking 1 seconds
BITMEX<key3> has 1322821 lines of records, taking 1 seconds
BITMEX<key4> has 2178846 lines of records, taking 2 seconds
BITMEX<key1> has 2377136 lines of records, taking 2 seconds
BITMEX<key7> has 1282649 lines of records, taking 1 seconds
BITMEX<key9> has 2322173 lines of records, taking 3 seconds
BITMEX<key6> has 3151901 lines of records, taking 3 seconds
BITMEX<key8> has 5223906 lines of records, taking 5 seconds
BITMEX<key11> has 14672360 lines of records, taking 14 seconds
BITMEX<key18> has 13753659 lines of records, taking 13 seconds
BITMEX<key20> has 17142423 lines of records, taking 17 seconds
BITMEX<key13> has 24016635 lines of records, taking 23 seconds
BITMEX<key12> has 34486869 lines of records, taking 32 seconds
till I encounter a key with huge data. And Python throw an error
Process Process-15:
Traceback (most recent call last):
File "/usr/lib/python3.7/multiprocessing/process.py", line 297, in _bootstrap
self.run()
File "/usr/lib/python3.7/multiprocessing/process.py", line 99, in run
self._target(*self._args, **self._kwargs)
File "<ipython-input-15-8b50676581a3>", line 8, in read_hdf5
ignore_index=True)
File "<string>", line 2, in __setitem__
File "/usr/lib/python3.7/multiprocessing/managers.py", line 795, in _callmethod
conn.send((self._id, methodname, args, kwds))
File "/usr/lib/python3.7/multiprocessing/connection.py", line 206, in send
self._send_bytes(_ForkingPickler.dumps(obj))
File "/usr/lib/python3.7/multiprocessing/connection.py", line 393, in _send_bytes
header = struct.pack("!i", n)
struct.error: 'i' format requires -2147483648 <= number <= 2147483647
OK. So it remains an open problem:
How to read into multiple data frame in parallel without wasting memory?